

Paradigm Shift Intervention Monitoring
 twitter
twitter

Commentary
Unchanged
Vaccine Targets Raise Pandemic Concerns
Recombinomics
Commentary 14:29
February 19, 2011
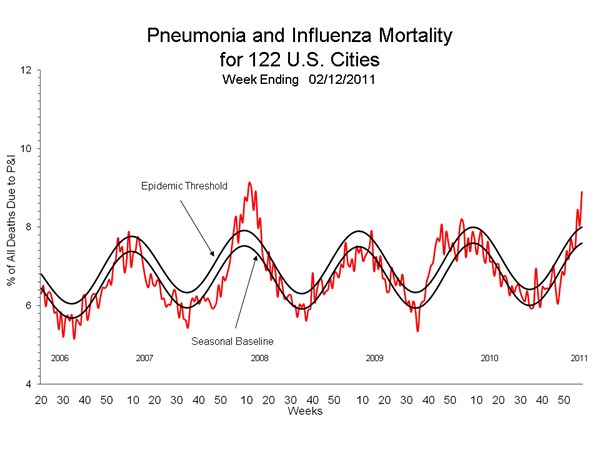
These record peaks are historically associated with vaccine mismatches. In the 2007/2008 season the three vaccine targets were A/Solomon Islands/3/2006, A/Wisconsin/67/2005, and B/Malaysia/2506/2004. All three targets for the following 2008/2009 season were changed, to A/Brisbane/59/2007, A/Brisbane/10/2007, B/Florida/4/2006.
The decision to leave the targets unchanged for the 2011/2012 was largely based on the antigenic characterization test, which has an extremely poor track record with regard to predictive value, sensitivity, or reproducibility. However, in spite of its abysmal performance, WHO consultants use this assay to recommend targets which are supplied to all major pharmaceutical companies making the flu vaccine, which are then distributed worldwide.
The recent history of the assay for seasonal and pandemic H1N1 has been reviewed recently here and here. The results for the 2007/2008 were particularly instructive. The ferret anti-sera used by the CDC could distinguish clade 1 (represented by New Caledonia/20/1999) from clade 2 (including Solomon Islands/03/2006), but did not distinguish between the three clade 2 sub-clades (Clade 2A - Solomon Islands/3/2006, Clade 2B - Brisbane/59/2007, and Clade 2C- Hong Kong/2562/2006). In the 2007/2008 season, the vaccine target, Solomon Island/3/2006 was no longer circulating. It had been replaced by the other two clade 2 sub-clades.
However, the CDC anti-sera was used to claim that Brisbane/59 and Hong Kong/2562 were a “good match”, which was refuted by the sequence data, the dominance of the clade 2B and clade 2C sub-clades, and a discriminating ferret anti-sera directed against Brisbane/59 grown in mammalian (MDCK) cells. As noted above, this target was then changed in the following northern hemisphere flu season. But the H1N1 mismatch for 2007/2008 season in the northern hemisphere, and 2008 in the southern hemisphere facilitated the emergence of Brisbane/59 like H1N1 which was Tamiflu resistant and had evolved away from the Brisbane/59 target, leading to widespread vaccine failure and the increase in Tamiflu resistance (H274Y) to 100% worldwide.
As noted earlier, the antigen characterization tests on 2009 and 2010 pandemic H1N1 isolates produced another series of irreproducible results including significant intra and inter-lab variations, including the absence of any H1N1 “low reactors” in the United States this season, in spite of reported 50% vaccine failure rate in the UK and record death rates as indicated by the latest P&I data in the graph above.
The latest recommendations by the WHO advisory committee raise serious concerns about the ability of these consultants to integrate clinical and sequence data with an extremely controversial and out dated antigen characterization test, which remains hazardous to the world’s health.
Media link
Recombinomics
Presentations
Recombinomics
Publications
Recombinomics
Paper
at Nature Precedings