

Paradigm Shift Intervention Monitoring
 twitter
twitter

Commentary
Spike
In H3N2 Low Reactors In The United States
Recombinomics
Commentary 18:25
March 24, 2011
Five hundred thirty-five (96.9%) of the 552 tested were characterized as A/Perth/16/2009-like, the influenza A (H3N2) component of the 2010-11 influenza vaccine for the Northern Hemisphere. Seventeen viruses (3.1%) of the 552 tested showed reduced titers with antiserum produced against A/Perth/16/2009.
The above comments from FluView weeks 9 and 10 respectively reflect the dramatic jump in H3N2 “low reactors” reported in the week 10 CDC FluView. Most of the sequences associated with the low reactor designation were reflected in HA sequences recently released by the CDC at GISAID. 14 of the released HA sequences were designated as low reactors by the CDC (see list here).
Phylogenetic analysis of the HA sequences raised serious questions about the sensitivity and reproducibility of the antigenic characterization test used to identify low reactors. This assay has been used for decades, but the release of sequences from tested isolates continues to raise serious questions about the validity of this assay.
This assay plays a pivotal role in the selection of new vaccine targets, as well as claims of “matches” between the vaccine target and circulating virus. These claims have a checkered and disturbing past, and the use of this assay to justify the use of this year’s targets in next year’s vaccine has raised significant concerns, as multiple reports describe vaccine failures associated with the current targets, and released sequences identify glaring inconsistencies.
The latest set of sequences extends the inconsistencies. Phylogenetic analysis identifies clustering of low reactors such as two isolates from Minnesota (A/Minnesota/17/2010 and A/Minnesota/18/2010) and one from Wyoming (A/Wyoming/09/2010). However, two other isolates from Minnesota (A/Minnesota/12/2010 and A/Minnesota/13/2010) and one from Maine (A/Maine/02/2010) are virtually identical to the three low reactors, but are not designated as low reactors. More dramatic examples were demonstrated by two closely related sequences from New Jersey (A/New Jersey/02/2010 and A/New Jersey/02/2011) which were designated as low reactors, while sequences which were identical to A/New Jersey/02/2011 (A/Alaska/01/2011, A/Delaware/10/2010, A/Nevada/04/2010, A/Pennsylvania/36/2010, A/Vermont/02/2011, A/Washington/04/2011) were not. Similarly, additional sequences closely related to the two New Jersey low reactors (A/Georgia/24/2010, A/Maine/02/2011, A/Louisiana/05/2010, A/Massachusetts/04/2010, A/New Jersey/01/2011, A/North Dakota/02/2011, A/Pennsylvania/41/2010, A/South Carolina/04/2010, A/South Dakota/02/2011), were also not designated as low reactors. The above sequences represent a few of the glaring inconsistencies, which are both common and widespread in the recently released sequences. Low reactors populate multiple additional branches, suggesting the vast majority of the recently released sequences are low reactors. Moreover, virtually all of the isolates were collected prior to the latest meetings of the WHO and US advisory committees on vaccine selection. Both committees recommended unchanged targets for the 2011/2012 influenza season in the northern hemisphere.
Additional data on vaccine failures in Air Force dependents as well as patients in Europe, including the UK, and Australia also revealed vaccine failures linked to the influenza A targets (A/California/07/2009 and A/Brisbane/16/2009, raising additional concerns over the recommendations to leave the targets unchanged. In addition, the pneumonia and influenza death rate has been at or above the epidemic threshold for the past seven weeks (tomorrow's week 11 report will show a jump to 8.61%), which is historically associated with vaccine mismatches such as this year and 2008, as seen in the graph below.
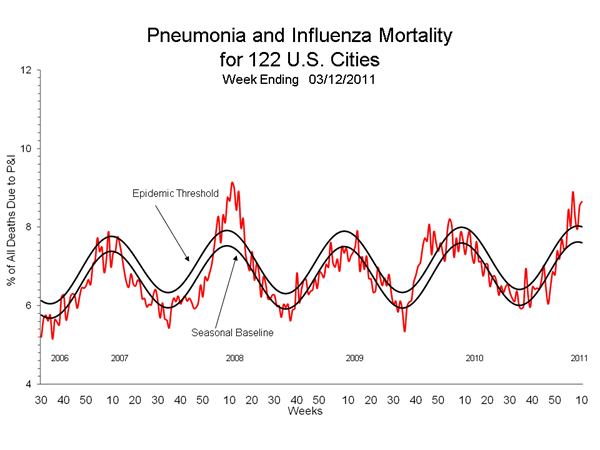
In 2008 all three vaccine targets were mismatched and changed for the following year, in contrast to this year when all three remained unchanged. Moreover, in 2008 the H1N1 target (A/Solomon Islands/3/2006) was initially called a match by the CDC, but phylogenetic analysis indicated that the target was no longer circulating and had been replaced by A/Brisbane/59/2007 and A/Hong Kong/2652/2006 and both prototype had a large number of HA changes, which were subsequently shown to represent antigenic differences easily identified by an anti-sera directed against the Brisbane target grown in mammalian cells, in marked contrast to the CDC anti-sera produce in chicken eggs which failed to distinguish the the sub-clades. This mismatch was associated with the widespread appearance of oseltamivir resistance (H274Y) in the 2007/2008 season in the northern hemisphere, and the fixing of H274Y during the 2008 season in the southern hemisphere. The switch to the Brisbane target in the 2008/2009 came after Brisbane/59 evolved further, leading to more vaccine failure. This year the vaccine mismatch linked to pandemic H1N1 is associated with the appearance of oseltamivir resistance in Delaware, raising concerns that the cluster signals the re-emergence of H274Y and possible fixing in the upcoming season in the southern hemisphere, where the mismatched vaccine is being distributed.
The continued reliance on the outdated, inconsistent, and insensitive antigenic characterization test by the WHO and US vaccine selection committees continues to be hazardous to the world’s health.
Media link
Recombinomics
Presentations
Recombinomics
Publications
Recombinomics
Paper
at Nature Precedings