

Paradigm Shift Intervention Monitoring
 twitter
twitter

Commentary
WHO Inconsistent Comments on Japan H274Y Tamiflu Resistance
Recombinomics
Commentary 16:40
June 19, 2011
The above comments from the latest WHO report on oseltamivir (Tamiflu) resiistance in H1N1 implies that the increasing frequency of Tamiflu resistance (H274Y) reported in Japan is due to Tamiflu treatment, but the latest sequences released by Japan’s National Institute on Infectious Diseases (NIID) indicate H274Y increases are due to community clonal expansion across the entire country.
NIID has recently released 43 NA sequences collected between Dec 2, 2010 and Feb 2, 2011. 13 of these sequences (largely from Jan collections) had H274Y and this high frequency (30.2%) may be markedly above the reported 1.5% due to selection of samples for sequencing, but the published sequences clearly demonstrate clonal expansion because 10 of the 13 sequences are virtually identical to each other and form a phylogenetic branch which is exclusively populated by isolates with H274Y, Moreover, as seen by the list (below) of these 10 isolates, they are from eight different prefectures, indicating the community clonal expansion has spread throughout the country. This spread is supported by the spike in the detection frequency (to 4.8% in Feb and 5.8% in Mar), which would again be linked to clonal expansion and not linked to Tamiflu treatment (Table 1):
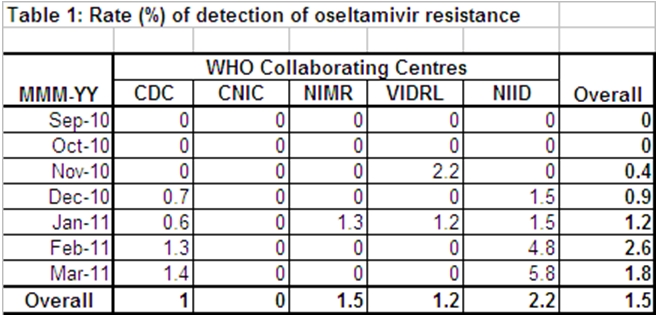
Thus, the basis for the WHO statement remains unclear. WHO consultants have agreed to report mixtures containing resistant sequences as wild type if the level of H274Y is below 50%, and therefore collection of samples after treatment may increase the level to more than 50%, leading to a resistance (H274Y) positive report. However, the virtual identity of the sequences from the 10 isolates below indicates the spread is via clonal expansion and not “spontaneous” mutations selected by Tamiflu treatment, as implied in the WHO statement above. Similarly, the wording of the WHO statement leaves open the possibility the patients were treated with Tamiflu after collection of the sample (which is common), and the classification of such patients as "Tamiflu treated" just adds to the confusion created by the “agreement” by consultants to not report Tamiflu resistance in samples where H274Y is below 50%.
In either case, the comments by WHO are grossly misleading and are little more than propaganda masquerading as science, creating the illusion that the increasing frequency of H274Y in community cases is linked to spontaneous mutations selected by Tamiflu use, rather than clonal expansion of a dominant sub-clade with H274Y.
A WHO explanation of these misleading reports is long overdue.
Closely related H274Y positive sequences:
A/HIROSHIMA/17/2011
A/WAKAYAMA/23/2011
A/SAPPORO/19/2011
A/NIIGATA/51/2011
A/HOKKAIDO/12/2011
A/YOKOHAMA/29/2011
A/NIIGATA/62/2011
A/KOBE/537/2011
A/YAMAGUCHI/35/2010
A/HIROSHIMA/49/2011
Media Link
Recombinomics
Presentations
Recombinomics
Publications
Recombinomics
Paper
at Nature Precedings