

Paradigm Shift Intervention Monitoring
 twitter
twitter

Commentary
Absence of
Influenza A Subtyping Increases Pandemic Concerns
Recombinomics Commentary 19:11
September 16, 2011
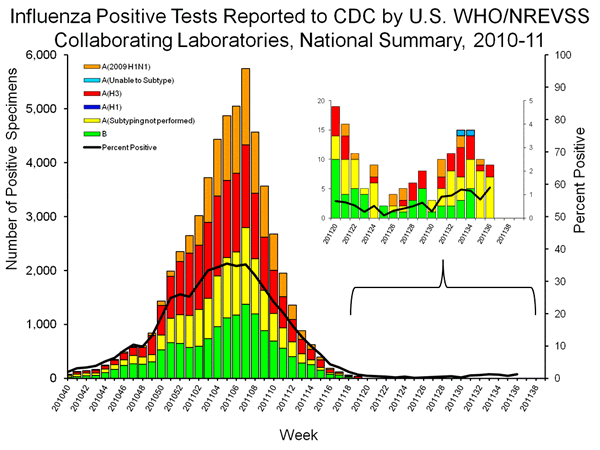
The recent detection of 4 trH3N2 cases has increased interest in the sub-type of influenza A positive samples because 2 or the 4 trH3N2 samples are list as unsubtypable as seen at the top of the bars representing week 33 and week 34. The absence of the other two cases from the graph is curious, since the H3 sequences from the four cases are virtually identical, and all four should have given an “unsubtypable” result. However, earlier trH3N2 had been classified as seasonal H3n2, raising concerns that the number of trH3N2 cases in the graph below is higher than the two designated for weeks 33 and 34.
The lack of sub-typing, as well as the absence of two of the four trH3N2 cases from the unsubtypable category raises concerns that the number of trH3N2 samples represented in the table below is markedly higher than the two unsubtypables or the four confirmed cases.
A recent paper on a trH1N1 from South Dakota described trH1N1 antibody detection in 40% of serum samples from students with swine contact. The M gene in the above isolate was closely related to the recent 2010 trH3N2 isolates, incluidng both from Pennsylvania (A/Pennsylvania/14 and A/Pennsylvania/40/2010). The recent trH3N2 isolates have swapped out the above M gene and replaced it with the M gene in pandemic H1N1 increasing tarnsmission efficiencies more. Thus, the four 2011 trH3N2 cases likely represent 100's or 1000's of human trH3N2 infections, raising serious surveillance concerns
The concerns are not reduced by the CDC sequences released for patients infected over the summer. There are only sequences from five patients and A/Maryland/17/2011 is the only patient which is not trH3N2.
An explanation for the lack of sub-typing as well as the two recent trH3N2 cases that are not listed as unsubtypable would be useful.
Recombinomics
Presentations
Recombinomics
Publications
Recombinomics
Paper
at Nature Precedings