

Paradigm Shift Intervention Monitoring
 twitter
twitter

Commentary
trH3N2
Unsubtypable Reporting Raises Pandemic Concerns
Recombinomics Commentary 19:00
October 7, 2011
The CDC has released full or partial sequences from the four recent cases (A/Indiana/08/2011, A/Pennsylvania/09/2011, A/Pennsylvania/10/2011, A/Pennsylvania/11/2011) at GISAID and Genbank and the sequences clearly indicate that all four isolates have the same constellation of genes including the acquisition on the M gene segment from pandemic H1N1. This acquisition is a concern because it was deemed “critical” for the jump of pandemic H1N1 from swine to humans, and raises concerns for efficient human transmission of H3N2. These concerns were increased by the identities between the four recent isolates from two distinct locations involving four independent infections, including the Indiana case (2M) who had no swine contact.
Detection of the trH3N2 cases is complicated by the H3 origin, which is human. Thus, an earlier trH3N2 case, A/Pennsylvania/40/2010 was initially reported as seasonal H3N2, but further analysis led to the determination that the infection was due to trH3N2, and the H3 sequence was closely related to other trH3N2 cases in 2010 (A/Wisconsin/12/2010 and A/Minnesota/11/2010) and presumably related to the trH3N2 that infected the daughter of the above Minnesota index case.
However, the 2011 trH3N2 sequences have evolved from the 2010 sequences and the underlying data for week 33 included an unsubtypable shortly after the early release of MMWR was made public. The underlying data from week 35 and 36 included unsubtypables, suggesting the additional H3 evolution led to a negative result for human H3 typing reagents. However, the underlying data (and accompanying figure) for week 37 and 38 removed the two unsubtypables, suggesting the cut-offs had been adjusted to classify the trH3N2 cases as seasonal H3N2 or to report such cases as negative (which might explain why only 2 of the 4 trH3N2 cases were reported as unsubtypables.
However, this week’s FluView increased the confusion because the underlying data again lists an unsubtypable for week 33 (which would correspond to the collection date for A/Pennsylvania/09/2011) and week 34 (which would correspond to collection dates for A/Pennsylvania/10/2011 or A/Pennsylvania/11/2011), but the figure (below) representing the underlying data only displays an unsubtypable for week 33.
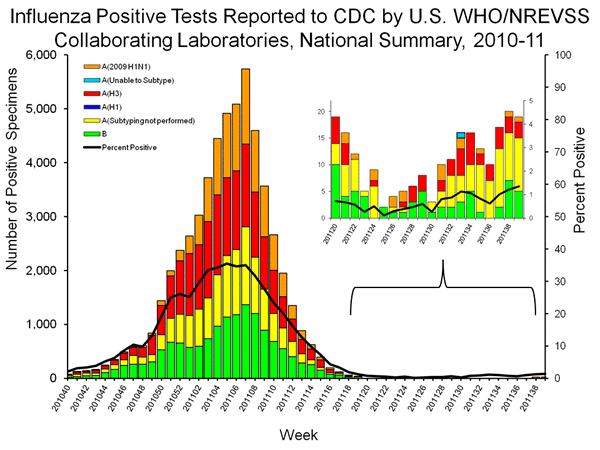
Thus, the absence of A/Indiana/08/2011 (samples collected in week 30), as well as one of the most recent confirmed cases from Pennsylvania (samples collected in week 34), and the inconsistencies for the reporting of unsubtypables in week 33 and 34 continue to raise concerns that the number of trH3N2 in the United States is markedly higher than those reported in the sub-typing table and figure reported in weekly FluView reports.
An explanation for these inconsistencies is overdue.
Recombinomics
Presentations
Recombinomics
Publications
Recombinomics
Paper
at Nature Precedings