

Paradigm Shift Intervention Monitoring
 twitter
twitter

Commentary
H3N2v
Transmission At 2011 Washington County Fair
Recombinomics
Commentary 20:00
October 11, 2012
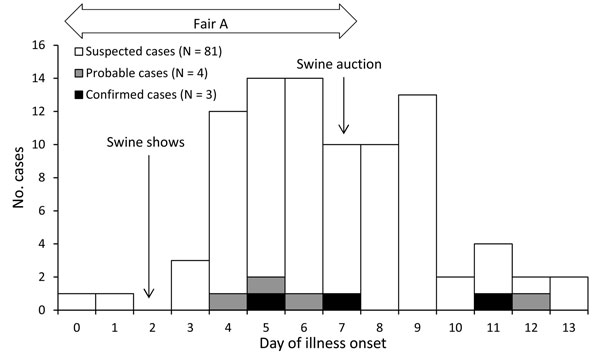
The CDC investigation identified four additional cases who were under the age of 4 and had H3N2v antibodies, which were classified as probable cases. Moreover, influenza-like illness was identified in 82 additional cases, which strongly support human to human transmission. Only one symptomatic swine was identified at the fair, and was removed in one day. Since the number of confirmed or suspect cases was high, and was due to multiple introductions, a swine source for these cases was unlikely.
However, the EID report focused on swine exposure, as seen in the above figure, and discounted swine exposure because the cases were identified at an agricultural fair. The CDC MMWR and subsequent reports centered on identification of additional cases without swine exposure, even though the case from Indiana had no sure exposure, and the number of suspect cases at the Washington County fair was high.
Subsequently, 2 cases were identified in Maine. These two cases attended another fair, but the time gap was too large for infection of the second case (A/Maine/07/2011) by the index case (A/Maine/06/2011) or through direct contacts at the fair. However, the sequences were virtually identical with each other, again supporting human to human transmission.
Three more cases were confirmed in Iowa. The three confirmed cases had no swine exposure and sequences were virtually identical. Two of the cases (A/Iowa/08/2011 and A/Iowa/09/2011) were classmates of the index case (A/Iowa/07/2011), while the father and brother of the index case were symptomatic, again signaling human to human transmission.
However, additional cases which matched these cases have not been identified in humans, but this sub-clade is now widespread in swine. Subsequent human cases have an N2 gene from a different lineage, with the exception of two cases from Michigan which have an N2 gene from the lineage seen in human 2011 cases, but have a PB1 gene with E618D, which was present in the six human H3N2v cases in 2010, as well as the first H3N2v case in the US, A/Kansas/13/2009.
Thus, there is little data to support extensive swine to human transmission, but that mode is targeted by the CDC, leading to a heavily biased sample set, as well as heavily biased analysis, as is evident in the upcoming paper.
Recombinomics
Presentations
Recombinomics
Publications
Recombinomics
Paper
at Nature Precedings