

Paradigm Shift Intervention Monitoring
 twitter
twitter

Commentary
Bizarre trH3N2
Unsubtypable CDC Reporting In FluView
Recombinomics Commentary 16:00
October 16, 2011
Four human infections with novel
influenza A viruses were detected in children from two states (Indiana
(1) and Pennsylvania (3)). All four children were infected with
swine-origin influenza A (H3N2) viruses; two were hospitalized; all
four have since recovered from their illness. The case in Indiana did
not report exposure to pigs prior to illness onset, although an
epidemiologic investigation concluded human to human transmission was
likely as a close contact reported direct contact with pigs prior to
the child’s illness onset. The three cases in Pennsylvania were
associated with attendance at a local fair where swine were exhibited.
The
above comments from the first CDC FluView
(week 40) for the 2011/2012 season note indirect linkage of the four
trH3N2 cases with swine, but acknowledge that there is no direct
linkage for the Indiana case, and is silent on direct contact for the
three Pennsylvania cases. Although the
three cases in Pennsylvania represent the largest number of confirmed
human trH3N2 from a single location, and the flu gene constellation
matches the Indiana case, the reporting of these cases in the
sub-typing figure and underlying data has been inconsistent and
confusing, raising serious questions about the status and reporting of
other trH3N2 cases.
The
inconsistencies in the 2011 trH3N2 cases follow delays in reporting
2010 cases. The clustering of 2010 cases
began a year ago and these clusters were announced in a WHO pager alert
for two cases, represented by A/Wisconsin/12/2010 and
A/Pennsylvania/14/2010 (PA/14/10). However,
at the time of the alert there was a second Pennsylvania case,
A/Pennsylvania/40/2010 (PA/40/10) under review, who was infected less
than a week prior to the Wisconsin case. However,
PA/40/10 was initially reported as seasonal H3N2, and the report of
this case as trH3N2 was delayed 5 months and reported in week 4 of 2011. Similarly, an Minnesota case (represented by
A/Minnesota/11/2010, was reported shortly after the pager alert, and
the index case was linked to additional symptomatic family members. The daughter of the index case subsequently
serologically lab confirmed to have been trH3N2, presumably by her
father or other family members since she had no reported linkage or
contact with swine. However, reporting of
this case was delayed 6 months, and testing of symptomatic family
members was reported as inconclusive. Moreover,
the CDC has used the isolate from the index case to develop a pandemic
trH3N2 vaccine.
Concern
due to the clustering of the 2010 trH3N2 cases was increased by the
clustering of the 2011 trH3N2 cases. All
four of the 2011 cases had the same constellation of flu genes. 5 of these genes were closely related to the
dominant trH3N2 sequences. The PB1 was
related to a prior triple reassortant outbreak at the Huron County fair
and represented by two human isolates, A/Ohio/01/2007 and
A/Ohio/02/2007. The NA gene in the 2011
isolates was closely related to the gene found in PA/14/10, while the
MP gene matched the sequences in pandemic H1N1. The
acquisition of the pandemic H1N1 MP gene increased concerns because the
MP gene was deemed critical for the jump of pandemic H1N1 from swine to
humans.
The
presence of this rare constellation of flu genes in all four human 2011
isolates, and absence of this constellation in any swine isolates,
strongly suggested that the 2011 trH3N2 was evolving and transmitting
in humans. The CDC claim of no sustained
transmission was blunted by the failure to identify a source for any of
the four confirmed cases. The Indian case
had no contact with swine, and although his caretaker did have contact,
neither the caretaker no the associated swine had flu symptoms, and no
evidence of swine influenza infections have been presented. Similarly, the three Washington County
attended the same agricultural fair where swine was exhibited, but no
symptoms have been reported for the swine at the fair and the sequences
of the cases indicates the isolate from the Schuylkill resident,
A/Pennsylvania/09/2011, were distinct from the other two cases,
signaling two source linked to the fair, but the sequences from the two
latter cases (A/Pennsylvania/10/2011 and A/Pennsylvania/11/2011) were
virtually identical to the Indiana case, A/Indiana/08/2011.
The
sequence data strongly suggests that the number of trH3N2 cases at the
fair was markedly higher than the three confirmed cases, but detection
of these cases is problematic because the H3 gene traces back to a
seasonal H3N2 infection of North American swine in the 1990’s. This origin led to the delay in reporting the
trH3N2 case, PA/40/10, which was initially reported as sesasonal H3N2. However, the 2011 H3 sequences have evolved
further, and the initial Washington County fair cases were reported as
unsubtypable, indicating this additional evolution had reduced the
sensitivity of the human H3 sub-typing reagents for the 2011 trH3N2
cases. Thus, samples identified as
influenza A positive, H3 negative would be likely trH3N2, which could
be confirmed by the CDC with a PCR test specific for trH3N2. This scenario is similar to initial testing of
pandemic H1n1, which were influenza A positive but H1 and H3 negative,
which were then sent to the CDC for further testing because the states
did not have the reagents for PCR testing for trH1N1.
When the volume of samples to the CDC overwhelmed the
testing, PCR kits for trH1N1 were then distributed to the states.
Currently,
the PCR tests for trH3N2 have not been distributed and detection is
dependent on states sending suspect cases to the CDC for further
analysis. However, the recent reports in
FluView have raised serious question about the state or CDC testing. The reports for PA/09/11 have been
inconsistent. All sequences from this case
have been made public at GISAID and Genbank, and the case was reported
in the early
release MMWR as well as the associated text in week 34 FluView, the
unsubtypable categorization first appeared in the weekend following
publication in these two source in the table of underlying data for the
sub-typing figure. The week 33 collection was
then reported in the table and figure in week
35 and week
36 reports, but was then absent in the week
37 and week
38 reports. It reappeared in the week
39 report, but as seen in the figure below,
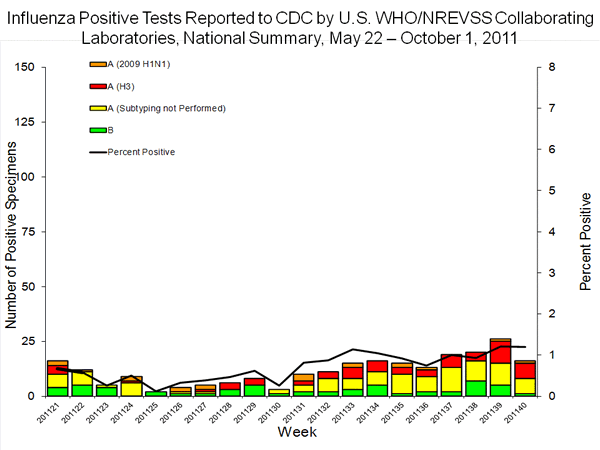
it again disappeared in the week 40 report, which had data for the first week of the 2011/2012 seasonal as well as the data for weeks 21-39, which included the collection dates for all four 2011 samples positive for trH3N2.

it again disappeared in the week 40 report, which had data for the first week of the 2011/2012 seasonal as well as the data for weeks 21-39, which included the collection dates for all four 2011 samples positive for trH3N2.
The
reporting of PA/10/11 was even more bizarre and inconsistent. The
unsubtypable (sample collected in week 34) was first reported in the
week 35 report and was again displayed in the week 36 report. However, it disappeared from the figure and
table in week 37 and week 38 reports, and then reappeared in the week
39 table, but not in the week 39 figure, which is based on the week 39
table.
Moreover,
the other two 2011 trH3N2 cases, from samples collected in week 30 and
week 34, have not been represented in any of the tables or figures (the
week 30 data has no reported unsubtypables or H3N2 cases).
The
inconsistent and bizarre reporting of the unsubtypables raises concerns
that these patterns are linked to the detection of additional trH3N2
cases or suspect cases, and associate changes in criteria for the
representation of these cases in the sub-typing figures and tables in
FluView.
These
changes have been made without explanation, and a clear definition of
criteria for reporting of unsubtypable / trH3N2 in the sub-typing
figure and table is long overdue.
Recombinomics
Presentations
Recombinomics
Publications
Recombinomics
Paper
at Nature Precedings