

Paradigm Shift Intervention Monitoring
 twitter
twitter

Commentary
Absence of
Maine trH3N2 Unsubtypable Increases Concerns
Recombinomics Commentary 12:45
November 2, 2011
Testing performed at Maine’s Health and Environmental Testing Laboratory on October 14, 2011 indicated a likely swine origin influenza A (H3N2) virus and this result was confirmed at CDC on October 16, 2011.
The above comments from the Maine CDC and the US CDC describe the results and dates for the first Maine unsubtypable, A/Maine/06/2011. The unsubtypale result was on October 14 in week 41. CDC confirmed that the unsubtypable (unable to sub-type or “inconclusive during subtyping”) was trH3N2 on October 16 (week 42) and the full sequence was released at GISAID on October 17 from a sample collected on October 10 (week 41).
Thus, although the key data points and characterization of this case were in week 41 and 42, the week 42 FluView has no unsubtypables listed in its figure for the 2011/2012 season (beginning with week 40) as seen below, and it is also not listed in the table representing the data in figure. Thus, there have been 5 trH3N2 cases confirmed in 2011 prior to week 42, but only two are listed in the subtyping table for the 2010/2011 season (A/Pennsylvania/09/2011 is listed in week 33, and the unsubtypable in week 34 is one of the latter isolates from Pennsylvania (A/Pennsylvania/10/2011 or A/Pennsylvania/11/2011).
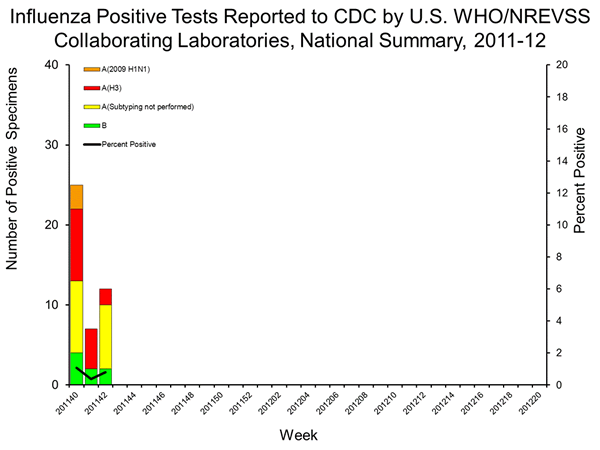
Moreover, this week the CDC released sequences from two additional trH3N2 cases (8M - A/Maine/07/2011 and 59M – A/Indiana/11/2011) which represent additional examples of this sub-clade in humans in 2011, which are identified by state labs due to the inability to sub-type with reagents directed against seasonal H3 or pandemic H1, suggesting these unsubtypables samples sent to the CDC will continue to increase, but most of these samples will not be included in the FluView subtyping tables.
Similarly, all described trH3N2 cases have some type of suspect swine “exposure” based on epidemiological studies, which is strongly contradicted by the absence of this sub-clade in swine. These patterns strongly suggest that data on trH3N2 cases with no swine “exposure” are being withheld by the CDC, pending epidemiological data.
Details on the trH3N2 cases with no swine exposure are long overdue, and should be released immediately.
Recombinomics
Presentations
Recombinomics
Publications
Recombinomics
Paper
at Nature Precedings